mRNA Display is a technology for creating ultrahigh diversity peptide and protein libraries (trillions of independent sequences!) for in vitro selection and directed evolution. Using mRNA display it is possible to select and evolve polypeptides with remarkable functional properties such as binding Proteins, RNA, Posttranslational Modifications, and Small Molecules.
How it Works
In mRNA display, mRNA molecules bearing a pendant 3’ puromycin are translated in vitro to generate covalent mRNA-protein fusions. In order to perform directed protein evolution experiments, synthesis of the mRNA-protein fusions is incorporated into an in vitro genetic cycle. An initial library is created in the form of double stranded linear DNA using PCR. Diversity in the library is generated using randomized DNA cassettes, doped cassettes, mutagenic PCR, or a combination of these methods. In vitro translation results in protein sequences covalently attached to their own mRNA. These mRNA-fusion molecules will then act as a molecular Rosetta’s Stone, and can be recovered after affinity maturation and amplified. Traditionally, mRNA display has been used in order to generate peptide and protein ligands which can be used as therapeutic or diagnostic reagents.
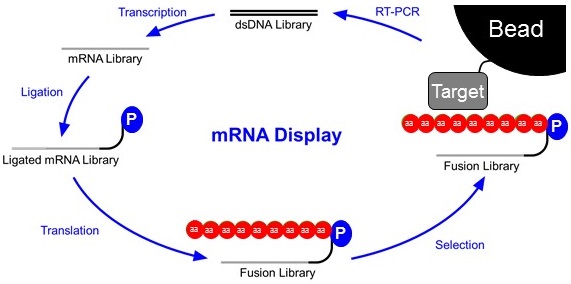
Libraries are constructed as double-stranded DNA (top). This DNA library will contain randomized stretches of nucleotides so that thousands to trillions of different proteins can be synthesized later in the cycle. This library is first transcribed into an mRNA library and then ligated to a short piece of DNA with a Puromycin connected to it’s 3’end. The ligated mRNA Library is then translated in vitro. As ribosomes read RNA and not DNA, the ribosome stalls at the mRNA-DNA interface. This allows time for the puromycin to enter the A site of the ribosome and form a covalent link between the transcript and the displayed peptide. This Fusion Library is then subjected to a selection step. Often times this is a co-immunoprecipitation of the fusion library against a target attached to a bead. This step is intended to wash away proteins with poor affinity for the target. After selection, the fusions that stuck to the bead are reverse transcribed into a new DNA library that will serve as the basis for the following round of selection.
Why mRNA Display?
mRNA Display has several significant advantages over other display techniques as a peptide and protein design tool:
1) higher diversity
2) lack of context dependence
3) strictly monomeric libraries
4) the ability to use an expanded genetic code
5) ease of use with high throughput sequence analysis.
With a diversity ~10,000 times greater than the primary immune repertoire, it is straightforward to discover much better ligands than with other methods such as phage display.