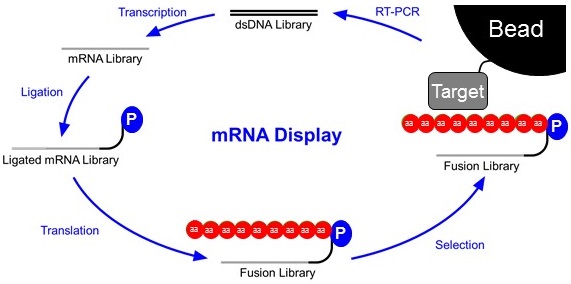
Research in the Roberts Lab involves engineering new peptides and proteins for biology, diagnosis, and therapy using mRNA display, invented by Richard Roberts. This method enables us to generate trillion-member libraries for in vitro selection and directed evolution experiments to discover ligands targeting proteins, nucleic acids, and small molecules. We are grateful for the Financial Support provided by federal agencies, corporations, private foundations, and charitable giving.
Our laboratory is joint between the Viterbi School of Engineering and the Dornsife College of Letters Arts and Sciences. Prof. Roberts holds appointments in the Mork Family Department of Chemical Engineering and Materials Science, the Department of Chemistry, and the Department of Molecular and Computational Biology and is a full member of the Norris Comprehensive Cancer Center.
Prof. Roberts also directs the USC Center for Peptide and Protein Engineering.